Interfacing with the DOSY Toolbox
There is no better way to introduce the DOSY toolbox than to cite its own web site.
The importance of High Resolution PFG-NMR data for mixture analysis is steadily increasing but there is no single way to process such data.
While there are a number of available methods, the commonest method is known as DOSY (diffusion-ordered spectroscopy),
and therefore it has become practice to refer to these data as DOSY data.
The major manufactures offers different implementations of DOSY processing (with a limited selection) in their software.
The DOSY Toolbox is a completely free programme that aims to incorporate many of the important processing schemes.
The DOSY Toolbox has a graphical user interface to make it easy to access a variety
of different processing schemes (and a command mode for more advanced options).
It is written in MATLAB, but can also be obtained as free standing compiled version that does not require a MATLAB installation.
The DOSY Toolbox is released as freeware under the GNU GPL license. It has also been described in a publication (see reference at the bottom of this page).
While the DOSY Toolbox is a stand-alone package capable of processing raw data (coming from the most common instruments) and create publication-ready plots, many users may prefer to use iNMR for generic tasks (like phase and baseline corrections, displaying, printing, etc.) and to employ the DOSY Toolbox for the diffusion-specific processing only. After a great collaborative effort between us and prof. Mathias Nilsson, we have provided the two programs with specific outlets that permit data transfer in both directions. The iNMR part of this bridge relies on external scripts, that can be easily examined and edited by any user. The spectra are exported as text (in a format called “DOSY Toolbox ASCII”). The format is simple by design, therefore any scriptable NMR software can be easily interfaced with the Toolbox. There is nothing specific to iNMR.
Here we describe in details the steps required to use the Toolbox in combination with iNMR.
Installation of the Toolbox
You need at least version 0.8 Release Candidate 3 (March 2010) or something more recent. If you haven't got the commercial program MATLAB, then you also need the MATLAB runtime (free). Both packages can be downloaded from the main site, which also includes generic installation instructions. When you have installed the runtime, it goes into this location:
/Applications/MATLAB/MATLAB_Compiler_Runtime/v710
It is important to know it, because it is a required argument to launch the DOSY Toolbox. The latter can be conveniently installed into your home directory or a sub-directory. The Toolbox comes as a folder. Into this folder there is a unix shell script that runs the program. The name of the script is something like: “run_DOSYToolbox.sh” In practice, to start the program you need to open the Terminal application and type a command structured in this way:
/path/to/the/script /path/to/the/runtime
For example:
DOSY_Toolbox/run_DOSYToolbox.sh /Applications/MATLAB/MATLAB_Compiler_Runtime/v710
Given that this command is quite long, it is convenient to save it too as another script, with a compact name like “DOSY.sh”.
The Terminal application remains important during the execution too. If the Toolbox detects an error, you will hear a beep and will see no message of explanation. In this case, look into the Terminal window, because the message has been written there.
From iNMR to the DOSY Toolbox
On the DOSY Toolbox web site you can also find a collection of test data. You can use it to test your installation and learn using the program.
Perform the standard processing with iNMR. After correcting the baseline, use a script to export the matrix. There are two different scripts: one for Varian data and another one for Bruker data. This happens because iNMR does not contain all the information required by the DOSY Toolbox and the scripts are forced to retrieve it from the original files, which are stored in different formats, as everybody knows.
The two scripts are quite similar and save the exported matrix into the same folder of the original data set, under the name “dosy.txt”. You can easily edit the scripts if you prefer a different destination or another name. Do not close the iNMR document, because it will be necessary when reimporting the processed DOSY matrix. Open the DOSY Toolbox and select the command: DOSY Files > Import > DOSY Toolbox ASCII. Navigate into the next dialog to select the “dosy.txt” file just saved. At this point you have to wait a while, because MATLAB is not efficient in converting ASCII files to numbers. You can optionally rewrite the scripts in order to save the matrix in binary format, which can be read in much less time (though this has not been tested yet).
Final Processing
The Toolbox is so called because it offers many different mathematical methods to process a DOSY experiment. The purpose of the present article is, however, simply to show you how the data transfer works and how to test it. Initially only the first spectrum is visible. To examine it better, you can use the buttons under the title “Scale” to adjust the intensity scale.
Click the buttons in the picture below in the order given by the numbers. Initially only the first row of buttons is visible. The other ones appear after you have selected a specific method (in our example: DOSY). After clicking the second button (Thresh), you should also click into the spectrum to specify the threshold (peaks under it are ignored by the program). To move the threshold to a different position, just click again. Once you have defined this threshold, you can start the proper processing with the button “Run”.
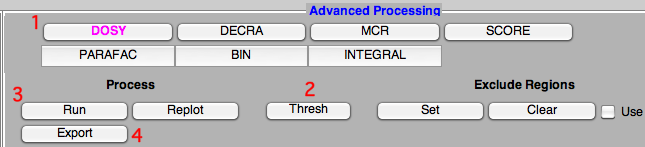
The resulting DOSY plot will appear into a new window. Return to the main window and click on “Export” to save the processed matrix.
From the DOSY Toolbox back into iNMR
Now return to the iNMR document. Basically, you need to paste the DOSY plot on it and save the result into a new file. Use this script that will do the whole work for you, including reopening the final document. Eventually you will have two windows open into iNMR: the starting data and the DOSY matrix. The latter is saved (by the script) into a folder called “from_DosyTb” and will appear next to the file you have exported from the Toolbox.
What else you can do
Both iNMR and the DOSY Toolbox are stand-alone packages which allow you to process a DOSY experiment from start to finish. Both are rapidly evolving programs, so you can expect new exciting possibilities to arise into the near future. You can extend iNMR by yourself by writing simple or complex scripts, like the ones we have used here. If you have MATLAB, you can also extend the capabilities of the DOSY Toolbox: its source is open. The program has not yet arrived at version 1, but it already benefits from the acknowledged experience of the author into the field of PFG-NMR. You should consult the scientific literature to learn more about the several methods proposed: HR-DOSY, DECRA, MCR and SCORE.
It is great to have the possibility of moving data back and forth between the two programs: now you can exploit each one for what it's best at. At the time of this writing, the documentation of the DOSY Toolbox (organized as a wiki) is rather laconic, which is positive if you already are a DOSY expert. Hopefully in the future there will be more information for the sake of the neophyte. We want to thank prof. Mathias Nilsson for all his efforts, in particular for the creation of the bridge described by this article. It really took a lot time and patience. If anybody is going to use his program, the reference to cite is:
M. Nilsson, The DOSY Toolbox: A new tool for processing PFG NMR diffusion data
Journal of Magnetic Resonance 2009, 200, 296-302.
DOI